Although zebrafish have been described numerous times as ideal organism models, manual handling of zebrafish embryos can turn out to be complex and time consuming due to their small size and the large quantities of samples usually required in an experimental setup (1). To increase efficiency of such studies, it has been crucial to develop fast and automated screening methods.
In this article we will discuss various high-throughput screening methods and automation technologies which have been developed and proven to be very helpful for zebrafish embryo and larval assays.
What is high-throughput screening ?
Today more than ever, the competition between pharmaceutical industries for drug discovery, development and sales is making it extremely important to have a fast, cost effective and efficacious way to screen a large number of compounds in a short amount of time.
High-throughput screening (HTS) is an effective way of running millions of chemical and biological screens simultaneously, using robotics and automation. Liquid handling devices, robotics, plate readers and software programs for instrument control and data processing are combined for fast and effective screening of biological samples. Biological samples include all biological levels, meaning molecules, cells, tissues, physiological pathways and model organisms (2).
HTS is very useful for drug screening by testing biological and biochemical activities. Additionally, the analysis of biological relevant compounds and targets is considerably accelerated compared to non-automated processes. Up to 103-106 small molecules of known structure can be screened in parallel, making it possible to analyse over 100’000 samples in a day (2). These numbers obviously depend on the type of assay and screen being done. HTS require a few basic elements (3):
Microplate preparation
Samples need to be prepared and arrayed in microplates. The choice of microplate will depend on the sample and assay being performed.
Configuration of the robotic workstation
A robotic workstation needs to be configured for automated handling of plates and data acquisition.
Choice of automation method
Setting up the most accurate assay and having an effective quality control system are critical for collecting high quality data with a clear distinction between negative and positive controls.
Data acquisition and analysis
Data needs to be acquired and processed which is commonly done by optical measurements.
Although HTS was initially developed for biochemical screens, nowadays it is also commonly used for other biological functions using multi-component biochemical assays. Cell-based assays of complex biological pathways have also been developed. The latest innovation in the field is whole model organisms screening, using for instance zebrafish (2).
Zebrafish are indeed excellent models for whole animal drug screening with excellent throughput. They are widely recognised as an ideal model for HTS chemical screening as they are closer to mammals from an evolutionary point of view than worms or flies. They also have a primary organ system very similar to humans. Additionally, they are cost effective and have a high fecundity rate, making a large number of embryos available for drug screening (4).
Using HTS and automation for zebrafish assays
Typical studies ran on zebrafish are phenotypic screens. As embryos are transparent, real time in vivo monitoring of morphological defects and effects on larval organ development allow identification of physiology altering agents in a whole organism (4).
Up until today, the sequential steps involved in such assays have been performed manually and are summarised in the figure below. These tasks can be tricky and time-consuming. As an example, testing one compound requires about 1000 embryos and each step needs to be carried out on each individual, requiring several hours. For trained personnel, manual egg collection, sorting, and distribution can take over an hour and if injections need to be done at the one cell stage, another 2 hours can be added to that time. Sorting and plating according to fluorescence can also take up to 2 hours. Finally, cell injection into larvae and larvae selection can respectively take up to 12 and 14 hours. Thus, testing just a single compound manually with zebrafish can take over 30 hours.
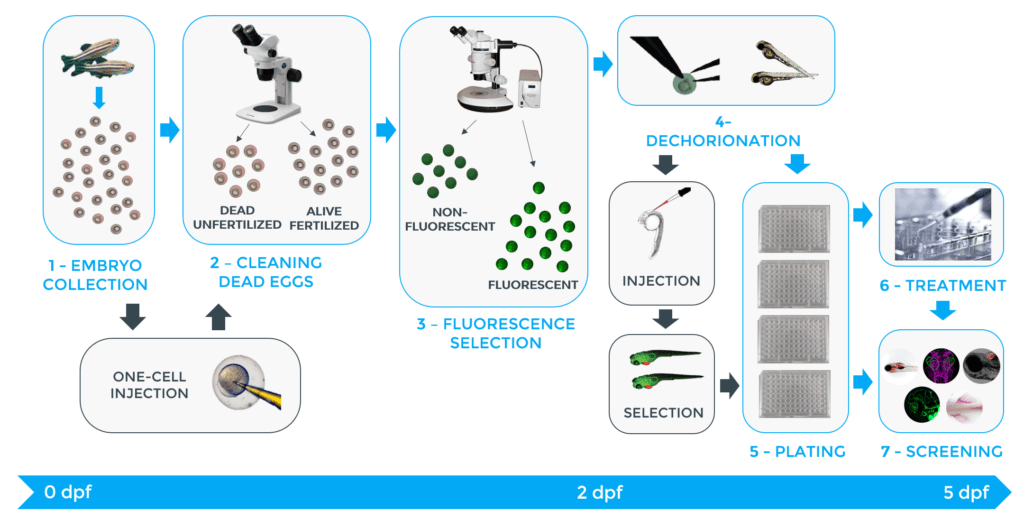
Figure 1: Manual steps of zebrafish embryo assays.
To increase efficiency of chemical and phenotypic screens conducted on zebrafish eggs, embryos or larvae, several companies have come up with ideas and developed devices to automate these procedures. To begin with, individual steps were automated, making one step among the whole procedure easier and faster. For instance, automated microscopy of zebrafish embryo and larvae is pretty advanced. Several automatic micro-injection systems have also been developed.
Anyhow, having a device capable of performing several of the sequential operations would further facilitate work with zebrafish. Several academic institutions are working on different prototypes. For instance, CSEM has developed an automatic cell-handling system which has sorting, injecting and feeding properties, facilitating handling of small biological entities such as zebrafish eggs and Xenopus oocytes (5). Another example is Fraunhofer which uses a machine learning algorithm for zebrafish egg sorting. Their final goal is to develop a high-throughput analysis system which can be integrated into a fully automated device to increase the rate of zebrafish egg processing (6). One last example which is worth mentioning is the BioRobotLab (KIT). They have also been working on a robotic device for automatic sorting of zebrafish eggs and larvae (7).
The Eggsorter by Bionomous
Bionomous has realised the necessity to find an effective solution to increase rapidity and facilitate zebrafish assays. The EggSorter has been designed to automatically screen, sort and dispense small biological entities, such as zebrafish eggs. Its main feature is a rotary wheel which can grab individual eggs and place them in front of an optic for analysis. A picture is taken and the data is examined using various algorithms for egg classification and sorting into multiple containers according to predefined optical features.
Several algorithms have been developed to sort eggs according to criteria such as fertilisation, reporter gene expression (GFP and mCherry for instance), stage of development or anomalies occurring during development. These algorithms can also be adapted to the experimental design.
The device is capable of sorting and processing one egg every second and is capable of handling biological entities up to 1.4 mm. Additionally, up to 36 samples can be evaluated simultaneously, increasing the efficiency of egg sorting. The results achieved following the sorting process are very encouraging: over 98.5% of the eggs survive the process and false discovery rate is lower than 4%.
Some other advantages of the EggSorter are that all of the processing is embedded in the device, thus no external computer is required and all of the configurations can directly be done on the EggSorter using a tactile screen or an android app. Besides, the device has a convenient size making it portable and easily movable in the lab. Finally, it is an affordable solution for increasing screening rate in laboratories working with large numbers of zebrafish embryos.
The numbers and information mentioned above should help illustrate the benefits of using the EggSorter while working with zebrafish. Additional numbers and information as well as a full description of the EggSorter is available on our website.
References
(1) Breitwieser, H., Dickmeis, T., Vogt, M., Ferg, M., & Pylatiuk, C. (2018). Fully Automated Pipetting Sorting System for Different Morphological Phenotypes of Zebrafish Embryos. SLAS technology, 23(2), 128–133. https://doi.org/10.1177/2472630317745780
(2) Wexler, P. (2014). High throughput screening. In Encyclopedia of toxicology. Amsterdam: Academic Press.
(3) High-throughput screening (HTS). (BMG LABTECH). Retrieved October 2nd, 2020, from https://www.bmglabtech.com/high-throughput-screening/
(4) Schutera, M., Dickmeis, T., Mione, M., Peravali, R., Marcato, D., Reischl, M., Mikut, R., & Pylatiuk, C. (2016). Automated phenotype pattern recognition of zebrafish for high-throughput screening. Bioengineered, 7(4), 261–265. https://doi.org/10.1080/21655979.2016.1197710
(5) Automated preprocessing of large microbiological entities. (CSEM). Retrieved October 2nd, 2020, from https://www.csem.ch/page.aspx?pid=38000
(6) Machine learning algorithm automatically sorts zebrafish eggs. (Fraunhofer IPA). Retrieved October 2nd, 2020, from https://www.fraunhofer.de/en/press/research-news/2018/September/Machine-learning-algorithm-automatically-sorts-zebrafish-eggs.html
(7) Pfriem, A., Pylatiuk, C., Alshut, R., Ziegener, B., Schulz, S., & Bretthauer, G. (2012). A modular, low-cost robot for zebrafish handling. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference, 2012, 980–983. https://doi.org/10.1109/EMBC.2012.6346097