Over the past years, zebrafish have proven to be excellent model systems for a wide panel of experiments. We have discussed throughout our previous articles their advantages and disadvantages, how they could be incorporated to various research fields and different automation techniques which facilitate their handling in laboratories. Today we will discuss 4 other model organisms: Drosophila melanogaster, Xenopus laevis, Caenorhabditis elegans and Arabidopsis thaliana, all have in common their genetic similarity to humans, their ease of care and prolific breeding. While this last feature is particularly useful for studies requiring large batch sizes, it can also significantly slow down and complicate experimental procedures. Manual manipulations of small creatures is a time consuming and strenuous task. Thus, the demand and necessity for efficient automation technologies has been increasing over the past years.
In this short article, we will discuss why Drosophila melanogaster, Xenopus laevis, Caenorhabditis elegans and Arabidopsis thaliana have become popular research models and how research could benefit from automation technologies to accelerate and improve efficiency of experimental procedures.
Model organisms
Model organisms are non-human species which have been extensively studied to uncover fundamental biological and human disease processes. Such organisms are essential as they allow in depth understanding of metabolic and developmental pathways as well as molecular and genetic factors involved in disease without causing harm to actual humans and reducing ethical concerns. However, not any organism can be considered as a model system as they must respond to all nine of the following characteristics.
- Appropriateness as an analogue
- Transferability of information
- Genetic uniformity of organisms
- Background knowledge of biological properties
- Cost and availability
- Generalisability of the results
- Ease of an adaptability to experimental manipulation
- Ecological consequences
- Ethical implications
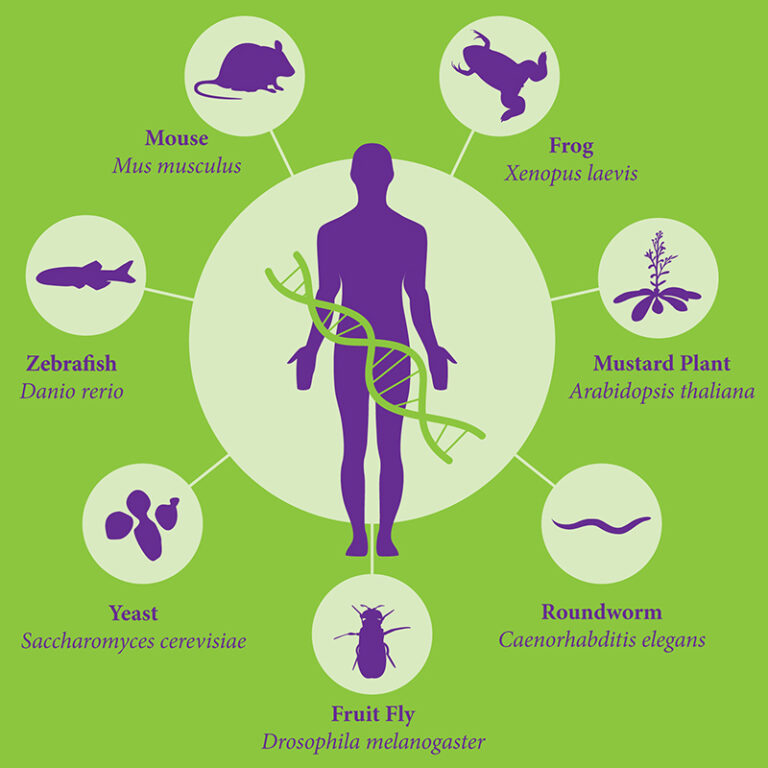
Figure 1: Model organisms are essential tools for biological research. A wide range of models are available for different types of experiments in various research fields1.
Therefore, an appropriate model system for each study should be chosen carefully. While Mus musculus is the most common model system for human disease research, other species have been found to be more adapted to other types of studies. The fruit fly Drosophila melanogaster for instance is the model of choice for development biology and human brain degenerative diseases. Xenopus laevis – the African clawed frog – is used to study embryonic development. Caenorhabditis elegans is also popular for development and differentiation studies. One last example we will discuss is Arabidopsis thaliana, a flowering plant widely used to study plant sciences such as genetics, evolution, population genetics and plant development.
Drosophila melanogaster
D. melanogaster is the common fruit fly you will find flying around your fruit in your kitchen. They are commonly used in research due to their rapid life cycle, their well-studied genetics and their high fecundity. One female can lay up to 100 eggs per day and 2000 in a lifetime2 and the offspring will develop in approximately 10 days. Therefore, several generations can be studied in only a few months. Moreover, D. melanogaster’s genome is small, well understood and well known. It only contains 139.5 megabase pairs over 4 pairs of chromosomes, 3 pairs of autosomes and one pair of sex chromosomes3. 60% of genes are conserved between the fruit fly and humans3 and 75% of human disease genes have a matching sequence in the fly4. They are popular genetic models in biomedical research for several human diseases, particularly neurodegenerative diseases, such as Parkinson’s, Huntington’s and Alzheimer’s disease5. Additionally, they are also suitable models to study aging, immunity, diabetes and cancer. While fruit flies were originally recognised as the model system of choice to study genetic patterns and inheritance, more recently they are also being used for environmental and mutagenesis studies.

Figure 2: Drosophila melanogaster.
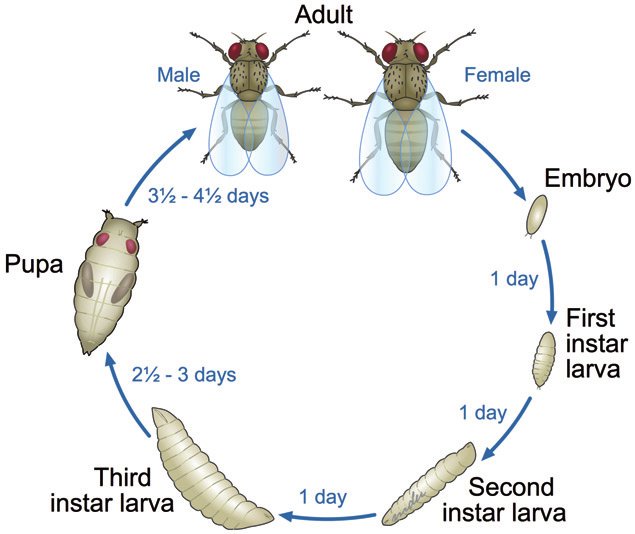
Figure 3: Timeline of Drosophila melanogaster development. The entire process takes about 10 days. The flies go through several stages of development before becoming mature adult flies6.
Xenopus Laevis
Xenopus laevis, also known as the African clawed frog, is an African aquatic frog originally found in Sub-Saharan Africa. They are highly adaptable and can survive and reproduce in a variety of conditions. When conditions aren’t optimal, particularly when the environment is too dry, they will bury themselves and remain dormant for up to a year. The African frog reproduces externally by fertilising eggs outside of the female’s body. In a laboratory setup, frogs can reproduce all year round following simple hormonal injections7. Several characteristics have made Xenopus an attractive model system in a variety of research fields. Not only are they cheap and easy to maintain, they are also highly prolific (they can produce thousands of eggs from each ovulation) and embryos are relatively large and easy to handle, which facilitates microsurgeries and injections of various substances such as proteins, nucleic acids or drugs7. Many key biological paradigms have been established thanks to Xenopus. For instance, Sir John Gurdon demonstrated using X. laevis that nuclei from differentiated cells could be reprogrammed following transplantation into enucleated eggs7. This experiment demonstrated how animal clones could be created. Additionally, Xenopus oocytes were used as an expression system to study biochemical and electrophysiological properties of ion and solute channels, carriers and transporters as well as signalling receptors8. Following these experiments, Xenopus have grown into a popular model system for molecular, cell and development biology of vertebrates. Nowadays, Xenopus can be used as a model to uncover underlying mechanisms of signal transduction, cell division, embryonic development, regenerative medicine and physiology. Moreover, Xenopus is gaining popularity for small molecule screens in the context of medical research and the development of novel therapies.

Figure 4: Xenopus leavis
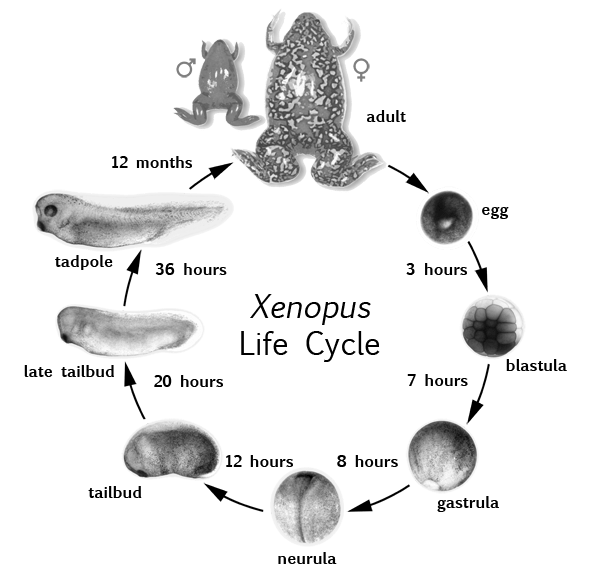
Figure 5: Xenopus leavis life cycle. Embryos develop externally. It only takes 36 hours for embryos to grow into a tadpole with a fully developed set of organs. However, it take 12 months for tadpoles to become mature adult frogs9.
Caenorhabditis elegans
C. elegans is a small transparent and unsegmented worm. It is the first multicellular organism to have had its entire genome sequenced and furthermore, its entire connectome described10. C. elegans has relatively short genome: it is made up of 100 million base pairs and only 6 pairs chromosomes. Moreover, most worms are hermaphrodite and do not have any sexual chromosomes. While these organisms remain fairly uncomplicated, they have a nervous system nonetheless. It is an elementary nervous system comprised of 32 neurones and which does not have any voltage gated channels or fire action potentials11. C. elegans was initially used in research for uncovering the mechanisms behind neural development. Nowadays, they are commonly used to study molecular mechanisms underlying behaviours such as learning, memory and mating, as well as molecular pathways behind metabolic diseases12. Additionally, their transparency makes them ideal to study cellular differentiation and development processes. Due to their small size (about 1 mm), they can easily be kept in a laboratory without any special equipment. Indeed, several hundred nematodes can be bred in a single agar plate on the appropriate growth medium13.
Figure 6: Caernorhabditis elegans14
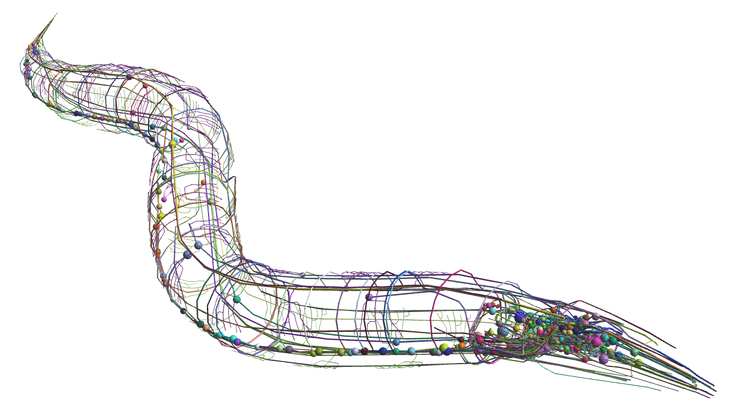
Figure 7: Caernorhabditis elegans connectome15.
Arabidopsis thaliana
Arabidopsis thaliana is a small flowering plant found in Europe and Asia. It is an annual plant and has a short life cycle. The central stem takes 3 weeks to grow and a fully-grown plant takes 6 weeks to develop. The plant can grow up to 25 cm long and the flowers are about 3 mm in diameter. The fruit is between 5 and 20 mm long and contains up to 30 seeds. A. thaliana self-pollinates. Therefore, A. thaliana can easily be grown in petri dishes in a typical laboratory seetup under fluorescent light to produce high quantities of seeds and specimens. Considering Arabidopsis is a eukaryote multicellular organism it has a relatively small genome with about 135 megabase pairs16. A. thaliana’s genome was the first plant genome to be sequenced and the first plant to be intensively investigated17. Up to date, over 3000 genes were uncovered and a large amount of data is available17. Therefore, Arabidopsis thaliana has grown into an excellent model system for understanding plant gene functions and uncovering pathways involved in plant physiology, biochemistry and development.
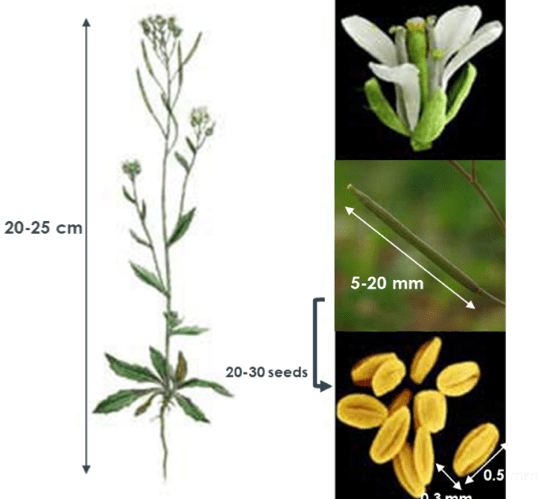
Figure 8: Arabidopsis thaliana. The whole plant can reach up to 25 cm long (left). The flower contains several fruits which can reach up to 20 mm and contain 25-30 seeds (right).

Figure 9: Arabidopsis thaliana can be grown in petri dishes in laboratories18.
Automation for small model organisms
Although the four discussed models have many advantages as animal models, they all have in common one disadvantage which can be an issue and slow down experimental procedures. The high fecundity of these models make them ideal for high-throughput screenings, meaning many individuals need to be handled within the context of a single study. However, their small sizes make experimental handling strenuous and time consuming. Procedures can include sorting, injections and imaging, among others. Automation technologies can significantly facilitate these tasks, not only by improving time efficiency but also by decreasing human errors. Automated phenotyping has already been developed and applied to several organisms over the past years. For instance, automated morphological phenotyping of drosophila using image analysis is widely employed19. However, this technique requires manual positioning of the flies and is not optimal. This has led to the development of improved techniques relying on body size or simultaneous morphological traits analysis. For Xenopus, automated microinjection systems for transfection protocols are available, reducing human labour time from one day to 4 hours20. Automation of data collection and analysis of C. elegans lifespan measurements in the context of aging experiments is also a trending topic with regards to novel automation technologies21. Finally, automated screenings of Arabidopsis seeds are also in demand. For instance, automated detection and sorting of germinated seeds directly into microplates has significantly improved efficiency of many multifactorial studies making use of large batches of seeds22. While our current automation technology has been thought and developed for zebrafish experimentation, we believe that we can adapt our device and extend its use to a wider panel of model organisms.
References
- Support Model Organism Databases! | SGD. https://www.yeastgenome.org/blog/support-model-organism-databases.
- Reeve, E. C. R. & Black, I. Encyclopedia of Genetics. (Taylor & Francis, 2001).
- Drosophila melanogaster (ID 47) – Genome – NCBI. https://www.ncbi.nlm.nih.gov/genome/?term=drosophila%20melanogaster.
- Reiter, L. T., Potocki, L., Chien, S., Gribskov, M. & Bier, E. A Systematic Analysis of Human Disease-Associated Gene Sequences In Drosophila melanogaster. Genome Res. 11, 1114–1125 (2001).
- Jaiswal, M., Sandoval, H., Zhang, K., Bayat, V. & Bellen, H. J. Probing Mechanisms That Underlie Human Neurodegenerative Diseases in Drosophila. Annu. Rev. Genet. 46, 371–396 (2012).
- Ong, C., Yung, L.-Y., Cai, Y., Bay, B.-H. & Baeg, G. Drosophila melanogaster as a model organism to study nanotoxicity. Nanotoxicology 9, 1–8 (2014).
- Ishibashi, S., Saldanha, F. Y. L. & Amaya, E. Xenopus as a Model Organism for Biomedical Research. in Basic Science Methods for Clinical Researchers 263–290 (Elsevier, 2017). doi:10.1016/B978-0-12-803077-6.00022-9.
- Schmitt, S. M., Gull, M. & Brändli, A. W. Engineering Xenopus embryos for phenotypic drug discovery screening. Adv. Drug Deliv. Rev. 69–70, 225–246 (2014).
- Introduction to Xenopus – Xenbase: The Xenopus Model Organism Knowledgebase. https://www.xenbase.org/anatomy/intro.do.
- Jabr, F. The Connectome Debate: Is Mapping the Mind of a Worm Worth It? Scientific Americanhttps://www.scientificamerican.com/article/c-elegans-connectome/.
- Kosinski, R. & Zaremba, M. Dynamics of the Model of the Caenorhabditis Elegans Neural Network. Acta Phys. Pol. B – ACTA PHYS POL B 38, (2007).
- Schafer, W. R. Deciphering the Neural and Molecular Mechanisms of C. elegans Behavior. Curr. Biol.15, R723–R729 (2005).
- Brenner, S. The Genetics of CAENORHABDITIS ELEGANS. Genetics 77, 71–94 (1974).
- The History of C. elegans. https://www.sibeliusnaturalproducts.com/the-history-of-c-elegans/.
- Neuroinformatics 2012: The NeuroML C. elegans Connectome. http://www.neuroinformatics2012.org/abstracts/the-neuroml-c.-elegans-connectome/index.html.
- TAIR – Genome Assembly. https://www.arabidopsis.org/portals/genAnnotation/gene_structural_annotation/agicomplete.jsp.
- Berardini, T. Z. et al. The Arabidopsis Information Resource: Making and Mining the ‘Gold Standard’ Annotated Reference Plant Genome. 18 (2016).
- CEA. Cultures d’Arabidopsis thaliana en milieu gélosé. CEA/Laboratoire Physiologie Cellulaire & Végétale https://www.lpcv.fr/Pages/Multimedia/Gal_A_thaliana.aspx (2019).
- Reeves, R. G. & Tautz, D. Automated Phenotyping Indicates Pupal Size in Drosophila Is a Highly Heritable Trait with an Apparent Polygenic Basis. G3 Genes Genomes Genet. 7, 1277–1286 (2017).
- Graf, S. F. et al. Fully Automated Microinjection System for Xenopus laevis Oocytes with Integrated Sorting and Collection. JALA J. Assoc. Lab. Autom. 16, 186–196 (2011).
- Felker, D. P., Robbins, C. E. & McCormick, M. A. Automation of C. elegans lifespan measurement. Transl. Med. Aging 4, 1–10 (2020).
- Colzani, M., Garbow, N., Berchet, V., Boix, M. & Lois, L. M. Automated Detection of Germinated Arabidopsis thaliana Seeds in Microplates. (2020).




