As described in last month’s article, New Approach Methodologies (NAMs) and Non-Animal Technologies (NATs), like organoids or fish eggs, are transforming the landscape of biomedical research, toxicology, and drug development. By offering human-relevant insights while reducing or replacing animal use, they hold the promise of more ethical, predictive, and cost-effective science. So, what’s limiting their scalability?
Introduction
Despite rapid advances, many NAMs and NATs remain difficult to scale. This is often due to manual processes, complex protocols, reliance on expert operators, and variability across labs. These factors limit reproducibility, standardization, and regulatory adoption, while driving up costs.
That’s where automation comes in. From improving reproducibility to cutting costs and saving time, it’s a key enabler for turning these promising tools into practical, scalable and standardized solutions. But developing such systems is no simple task, so how do we bridge the gap between promise and practice?
In this article, we dive into the critical role automation plays in making NAMs and NATs ready for the real world.
Sample handling
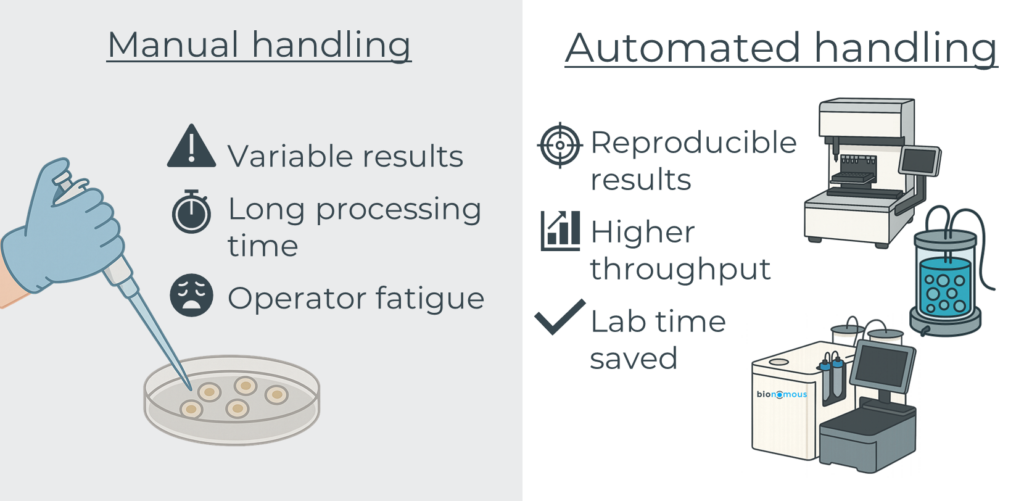
Sample handling is one of the most overlooked, yet critical, steps in any laboratory workflow, especially when working with NAMs and NATs. Whether it involves pipetting, dispensing, transferring, shaking, or simply moving samples between stages, these actions demand precision, care, and time.
This becomes even more complex when dealing with small, fragile biological models, such as organoids or fish eggs, produced in large quantities for high-throughput screening (HTS). Add to this the use of costly reagents and long cultivation times, and even a minor handling error can result in the loss of weeks of work and significant financial investment.
Automation offers powerful solutions to these challenges. Purpose-built systems can ensure:
- Gentle handling of sensitive biological materials
- Consistent and reproducible outcomes across large batches
- Significant time savings while reducing operator fatigue and repetitive strain
However, automating sample handling for NAMs is not without its challenges. Key concerns include maintaining sterility throughout the process, ensuring biocompatibility of materials, and integrating automated handling into broader workflows involving culture, treatment, and analysis. Furthermore, the initial investment in developing or adopting such systems can be substantial.
Nevertheless, automation is already making its way into sample handling through several commercially available systems, covering different steps of the sample handling workflow.
The first step in working with cells, spheroids, or organoids involves forming, culturing, and maturing these miniature biological systems. This process typically requires multiple liquid exchanges, temperature and gas control, application of various stimuli, and regular quality assessments. Several devices are already commercially available for 2D cell culture, especially for cell therapy (1), such as the Cocoon (Lonza), CliniMACS (Miltenyi), Celligent™ (Cell X Technologies), or C-Stem™ (TreeFrog Therapeutics), while many others are still at the R&D stage (e.g., LimeONE by Limula). For 3D cell culture systems, most protocols are still performed manually, but a few devices have started to appear on the market, like OrgaNest® (Celloid), CellXpress.ai (Molecular Devices), and ClinoStar® (CelVivo), or are close to market launch, such as the RPMotion bioreactor (Orgonex).
Automated liquid handlers are increasingly used to streamline 2D and 3D cell culture workflows. By automating tasks like media changes, reagent addition, and serial dilutions, these platforms reduce variability and boost throughput while ensuring consistency. In 2D cultures, they support applications from routine maintenance to high-throughput drug screening. For 3D models like spheroids and organoids, they facilitate gentle liquid handling to preserve structural integrity, though integration with bioreactors and imaging systems remains a challenge. Leading commercially available systems include the Microlab STAR (Hamilton), Fluent (Tecan Group), Flex (Opentrons), and Biomek (Beckman Coulter). Depending on the platform, these systems can offer adaptable, scalable solutions suitable for research, process development, or even production settings. While challenges such as large footprints, high costs, and protocol complexity remain, these systems represent a key building block in the move toward scalable, standardized, and automated workflows.
Automated sorting platforms help standardize biological sample selection by classifying cells or multicellular structures based on traits like morphology, viability, or developmental stage. This step is essential for ensuring experimental consistency, minimizing waste, and enhancing reproducibility. For 2D cultures, FACS (Fluorescence-Activated Cell Sorting) remains a benchmark method for precise, high-throughput sorting of individual cells. More recently, advanced systems like Cyto-Mine (Sphere bio) combine single-cell encapsulation, screening, and dispensing into an integrated microfluidic workflow, accelerating applications such as monoclonal antibody discovery or cell line development. In contrast, 3D models such as zebrafish eggs require gentler handling. Devices like our EggSorter use image-based AI to sort live eggs based on fertilization status or developmental stage, preserving integrity while reducing manual handling time. Together, these tools enable more scalable and standardized workflows while respecting the unique needs of each model system.
Overall, the growing ecosystem of automation platforms has laid a strong foundation for scalable, standardized workflows in cell and organoid handling. While challenges like integration complexity and cost remain, ongoing innovations are steadily bridging these gaps, with the outlook pointing toward fully closed systems.
Image analysis
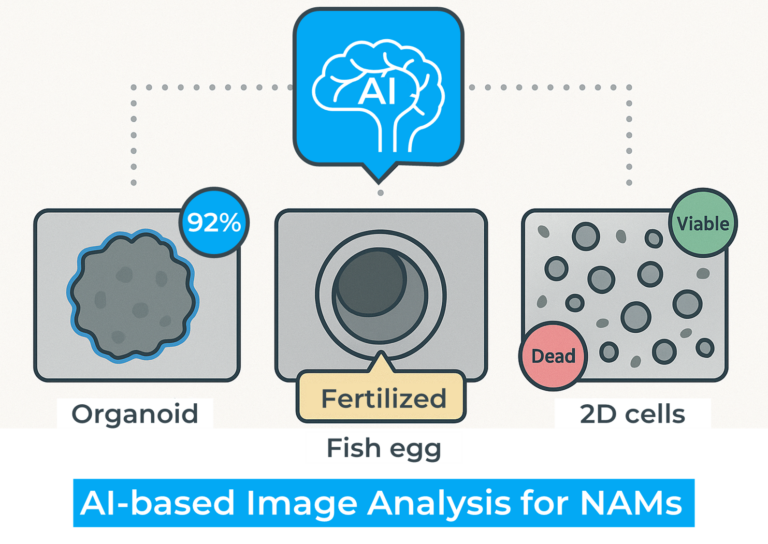
Microscopy-based image analysis plays a central role in evaluating biological samples in NAMs & NATs workflows: whether assessing viability in cells, organoids, or zebrafish eggs, or screening for drug and toxicological effects. However, manual analysis introduces variability, consumes time, and is often difficult to standardize. Unique morphologies, large batch sizes, and the time-sensitive nature of many models make consistency a persistent challenge.
Automating image analysis, particularly with artificial intelligence (AI), offers a transformative solution. It enables reproducible classification, reduces hands-on time, and captures expert decision-making in algorithmic form. AI can also track and store data automatically, making it easier for users to review and verify classifications.
A growing number of tools are emerging to support image analysis in NAMs and NATs. Major microscope and integrated imaging system providers like Nikon, Zeiss, Evident Scientific, Tecan, and Molecular Devices have developed high-performance instruments and dedicated software tailored for 3D cell culture, especially organoids. The american company, Ramona Optics, provides also different solutions for high resolution screening of samples in multi-well plates. These solutions offer powerful, ready-to-use pipelines, but often lack the flexibility for user-defined customization.
In parallel, deep learning (DL) tools have advanced rapidly. Open-source models like MOrgAna (morphological classification), deepOrganoid (viability assessment), OrgaExtractor (colonic organoid recognition), and 3DCellScope (3D reconstruction) empower researchers to build and adapt their own analysis workflows (2, 3). While these tools are highly customizable and promote broader understanding, further efforts are needed to harmonize analytical methods across laboratories.
For zebrafish eggs and similar small models, platforms like LabSwipe offer researchers the ability to train custom AI models using their own datasets and criteria, while maintaining full control over their data. Learn more on our dedicated page.
The field of AI-driven image analysis is progressing rapidly, offering powerful capabilities for complex biological models. However, developing robust algorithms still depends on large, high-quality annotated datasets, which remain scarce, especially for fragile or long-cultured systems like organoids. Ensuring that models generalize well across labs and experimental setups is another ongoing challenge.
Despite these hurdles, AI-powered image analysis stands to become a cornerstone of scalable, reproducible NAMs and NATs, bringing precision, flexibility, and speed to the next generation of model-based research.
Data recovery
In high-throughput NAMs and NATs workflows, acquiring high-quality data is only the first step. Equally critical is the ability to retrieve and organize this information efficiently. Without proper data recovery tools, valuable insights can be lost in fragmented files, inconsistent formats, or time-consuming manual extraction.
Automation plays a key role in addressing this issue. It allows data to be compiled into centralized, standardized files, saving time and making downstream analysis significantly more efficient. However, these benefits come with challenges. Ensuring data privacy, maintaining user control, and adapting to specific use cases remain key considerations when designing or choosing such systems.
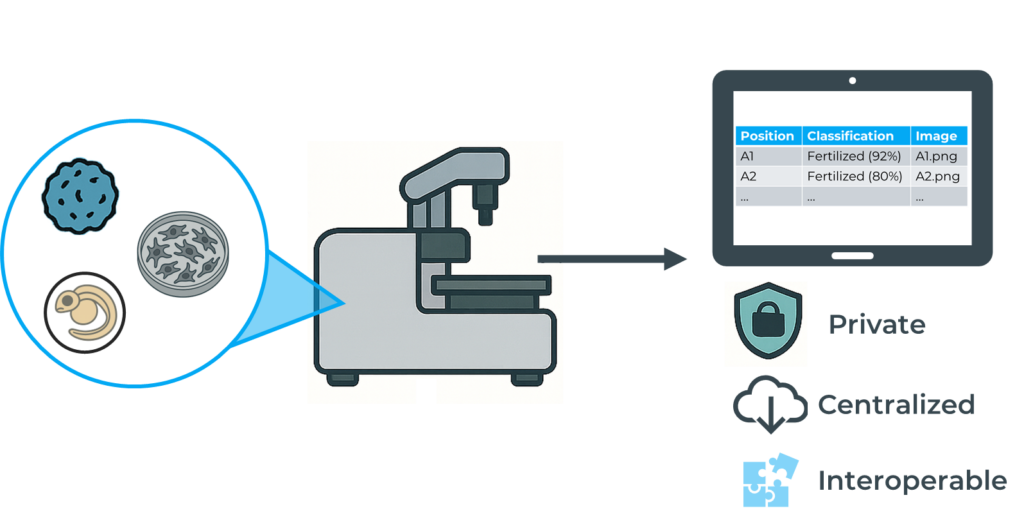
At Bionomous, for instance, we have developed a dedicated Android application that not only controls the device but also enables intuitive access to experiment data. The main output is a comprehensive CSV file summarizing key parameters such as the number of entities processed, classification results, image references, and output positions, all exportable and ready for further use.
As automated tools continue to evolve, integrating intelligent, accessible and reliable data recovery solutions will be essential to unlock the full potential of non-animal testing approaches.
Conclusions & outlooks
Automation is rapidly becoming an essential component across all stages of NAMs and NATs workflows: from the culture of biological models to their imaging, analysis, and data interpretation. As these methodologies scale up in relevance and adoption, automated systems offer unmatched advantages in standardization, reproducibility, and throughput.
Significant progress has been made on both the hardware and software fronts. On the hardware side, purpose-built devices for cell and organoid culture, automated sorting, and high-throughput imaging are becoming increasingly accessible. Simultaneously, advances in software, particularly in artificial intelligence, image analysis, and data centralization, are enabling more accurate, efficient, and traceable workflows. Together, these developments are shaping a new era of preclinical testing that aligns with ethical and scientific imperatives.
Looking ahead, the field must address one key challenge: aligning these systems with evolving regulatory frameworks. Regulatory acceptance will be particularly decisive, as institutions like the OECD, EMA, and FDA begin to set expectations for standardization, data quality, and traceability in non-animal testing approaches. If successfully addressed, automation will not only accelerate the transition away from animal testing but also help establish robust, integrated, and globally accepted NAMs platforms.
References
- Hwang, J. et al., (2024). Automation of cell culture and organ-on-chip platforms: Towards reproducibility and scalability in drug discovery. European Journal of Pharmaceutics and Biopharmaceutics. Advance online publication. https://doi.org/10.1016/j.jconrel.2025.02.057
- Rocha, M. C. et al., (2023). Organoid image analysis and machine learning-based classification in a label-free drug screening pipeline. Scientific Reports, 13, 19900. https://doi.org/10.1038/s41598-023-46485-2
- Ong, H. T. et al., (2025). Digitalized organoids: Integrated pipeline for high-speed 3D analysis of organoid structures using multilevel segmentation and cellular topology. Nature Methods, 22(6), 1343–1354. https://doi.org/10.1038/s41592-025-02685-4