Since 1901, Nobel Prizes have been awarded to “those who, during the preceding year, have conferred the greatest benefit to humankind”, in Alfred Nobel’s words1. Last year, Emmanuelle Charpentier and Jennifer Doudna were awarded with this recognized prize in the field of chemistry for their contributions to the genetic engineering tool known as CRISPR/Cas92.
CRISPR/Cas9 has been extensively studied and meticulously developed during these last years for efficient and precise genome editing. In this article, we will review the scientific paper: Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA3, published in Angewandte Chemie in March 2020. In this paper, the authors describe a modification of the CRISPR/Cas9 system so that it can be regulated in a precise manner by light, and they demonstrate its application in zebrafish.
Here we will give an overview of the CRISPR/Cas9 system, we will explain the modifications the authors of the paper made on that system, and we will go through the results they obtained in zebrafish.
CRISPR/Cas9: the gene editing system that is revolutionising 21st century science
CRISPR, short for clustered regularly interspaced short palindromic repeats, refers to a family of DNA sequences found in the genome of prokaryotic organisms such as bacteria and archaea4. This term was first used in the 1990s by Francisco Mojica who recognised for the first time these sequences in the genome of the archaeon Haloferax and stated that these DNA sequences that had been thought to be disparate repeat sequences actually shared a common set of features5. He also hypothesised that they may act as an adaptive immune system in these organisms, fact that was later on demonstrated by different research groups6.
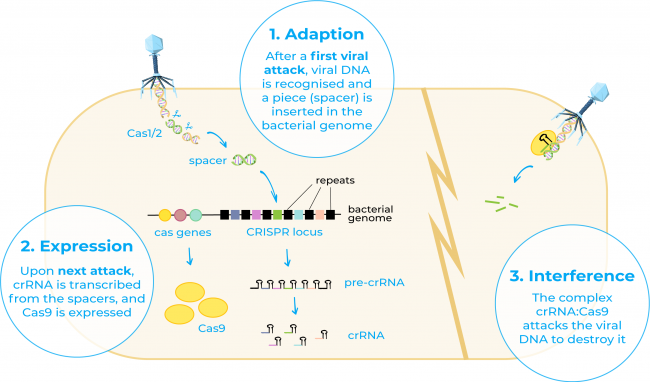
On the other hand, Cas9 or CRISPR-associated protein 9 is an enzyme that uses CRISPR sequences as a guide to recognise and cleave specific strands of DNA that are complementary to the CRISPR sequence. Cas9 enzyme together with CRISPR sequences form the basics of the technology CRISPR/Cas94.
Since the discovery of CRISPR and Cas 9 several studies began to fill in the details on exactly how CRISPR/Cas systems work. And it was in 2012 when Georges Church, Emmanuelle Charpentier Jennifer Doudna, and Feng Zhang isolated the components of the CRISPR-Cas9 system and adapted them to function as a tool to modify genomes7-9.
This programmable gene editing system has been extensively studied and developed since then and has been used in many applications in the fields of agriculture, basic science and medicine. In less than a decade, researchers have used CRISPR/Cas9 to develop genome-edited crops, insects and genetic models. Moreover, clinical trials are under way to use this tool to treat sickle-cell anaemia, hereditary blindness and cancer10.
Guide RNAs: targeting the gene of interest
There are two key molecules that allow the CRISPR/Cas9 system to work as a gene-editing tool: the enzyme Cas9 and a piece of RNA called guide RNA (gRNA)11. Cas 9 acts as a pair of ‘molecular scissors’ that can cut the DNA at a specific location, thanks to the guiding of the gRNA. The gRNA consists of a small piece of synthetic RNA composed of a scaffold sequence necessary to bind to Cas enzymes and a user-defined spacer sequence (of about 20 bases) that defines the genomic target of the Cas protein11.
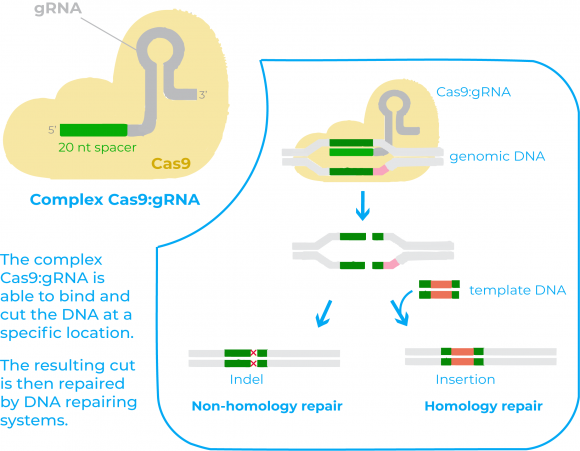
Once expressed together, the Cas9 protein and the gRNA from a complex that is able to bind to the target DNA and that allows Cas9 to cut it at a specific location11. The resulting cut in the DNA by Cas9 is then repaired by DNA repairing systems. After these repairing systems act it may happen two things: either the DNA is re-ligated leaving a scar in form of insertion/deletion (indel) mutations, or if there is a DNA molecule that can serve as a template for the repairing, the process will allow to insert a specific DNA sequence in the host genome11.
In this way, researchers have worked to modify these two components in many ways so that the genome editing is efficient, precise and in a customizable fashion.
Spatiotemporal control of CRISPR/Cas9
One of the challenges when aiming for higher genomic editing is to limit the window of CRISPR/Cas9 activity as well as to probe spatiotemporal controlled gene function. Efforts for the conditional controlling of CRISPR/Cas9 activity include regulation of Cas9 protein to restore its function upon external stimulation (for example, small-molecule-induced Cas9 activation or light activation of Cas9) or conditional control of chemically modified gRNA3.
Regarding the latter, several approaches have been described: protectors for gRNA activity, ligand-dependent RNA cleavage and deprotection, or small-molecule-induced reassembly of the Cas9:gRNA complex, among others3.
However, the authors of the paper defend that these systems are still limited by the requirement of first, protein engineering for Cas9 modifications, and secondly by the requirement of a third cellular component for the conditional controlling through gRNA modifications3.
To overcome the limitations to which they refer, the authors propose a new way of modifying the gRNA so that the interaction between the Cas9:gRNA complex and the DNA can be controlled by the use of light3.
A new light controlled CRISPR/Cas9 system
Light irradiation is an excellent external trigger to afford high-resolution control over biological processes, as irradiation can be precisely controlled in time, space and amplitude. In the referred paper, they design what is known as a photocaged gRNA.
The mechanism of photocaged RNAs is based on the installation of a photocleavable protecting group onto some of the bases of the RNA that renders it inactive (also known as ‘caging’). When this RNA is irradiated with non-damaging UV light, the caging group is removed, and the activity of the RNA is restored. This system has already been successfully applied for many biological functions where RNA is implicated, but not to control CRISPR/Cas9 activity12.
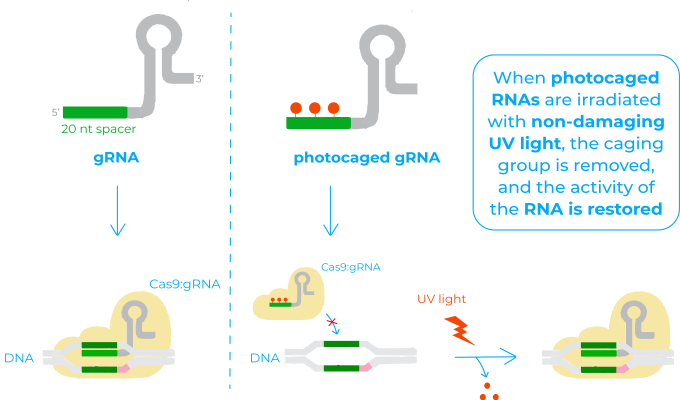
In this sense, the photocaged gRNA they use is designed so that the 20 nucleotides base-pairing region that recognises the target DNA has some caged bases. Therefore, before irradiation, this region is unable to bind to the DNA and Cas9 unable to act. Once irradiated with light, photolysis of the caged bases restores the native base-pairing of the gRNA. The target DNA is then recognised, the complex Cas9:gRNA binds to it and Cas9 cuts the DNA3.
Demonstration of photocaged gRNAs application in zebrafish
The Cas9:gRNA complex is an excellent tool for gene editing in zebrafish embryos, due to ease of assembly and injection into the fertilized oocyte13. Moreover, transparency of embryos allows for optical control.
Among the experiments done in the paper, one of them consisted in disrupting the expression of the fluorescent protein EGFP in a transgenic line of zebrafish. They injected a Cas9:gRNA complex with a caged gRNA targeting EGFP gene in the genome of that transgenic line. Hence, when the embryos were irradiated with UV light, they lost the EGFP expression3.
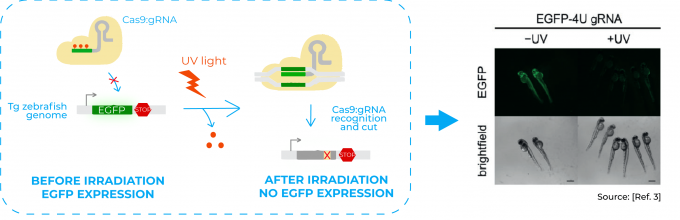
Besides, to show how light provides a unique opportunity to convey temporal control, they also targeted an endogenous gene in zebrafish important for the development of pigmentation in the embryos. They activated the Cas9:gRNA complex targeting this gene at different time points of zebrafish embryo development and showed that phenotypes changed accordingly3.
Conclusion
As an external trigger, light provides a unique opportunity to control gene editing with spatial and temporal resolution. Zebrafish embryos are an extraordinary model to demonstrate the potential of new gene editing tools based on the use of light, since they are transparent.
This new approach using photocaged gRNA offer exceptional features, including a rapid and non-invasive activation of CRISPR/Cas9 activity, precise spatial and temporal control and programmability of the light-activated gRNA sequence3. Thereby, this new technique can be used to investigate spatiotemporally complex physiological events since it will help to better understand the dynamics of gene regulation.
Shared if you liked it!
References
- Alfred Nobel’s Will. Nobel Prize. Retrieved in July 2021. https://www.nobelprize.org/alfred-nobel/alfred-nobels-will/
- Press release: The Nobel Prize in Chemistry 2020. Nobel Prize. Retrieved in July 2021. https://www.nobelprize.org/prizes/chemistry/2020/press-release/
- Zhou, W. et al. (2020) Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA. Angewandte Chemie (International ed. in English). https://doi.org/10.1002/anie.201914575
- Gupta, D. et al. (2019). CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life sciences. https://doi.org/10.1016/j.lfs.2019.116636
- Mojica, F. J., Juez, G., Rodríguez-Valera, F. (1993). Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Molecular microbiology. https://doi.org/10.1111/j.1365-2958.1993.tb01721.x
- Mojica, F. J. et al. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. https://doi.org/10.1007/s00239-004-0046-3
- Mali, P. et al. (2013). RNA-guided human genome engineering via Cas9. Science. https://doi.org/10.1126/science.1232033
- Cong, L. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. https://doi.org/10.1126/science.1231143
- Jinek, M. et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. https://doi.org/10.1126/science.1225829
- Hirakawa, M. P. et al. (2020). Gene editing and CRISPR in the clinic: current and future perspectives. Bioscience reports. https://doi.org/10.1042/BSR20200127
- Doudna, J. A., & Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. https://doi.org/10.1126/science.1258096
- Hemphill, J. et al. (2014). Site-specific promoter caging enables optochemical gene activation in cells and animals. Journal of the American Chemical Society. https://doi.org/10.1021/ja500327g
- Xin Y., Duan C. (2018) Microinjection of Antisense Morpholinos, CRISPR/Cas9 RNP, and RNA/DNA into Zebrafish Embryos. Methods in Molecular Biology. Humana Press. https://doi.org/10.1007/978-1-4939-7665-2_18