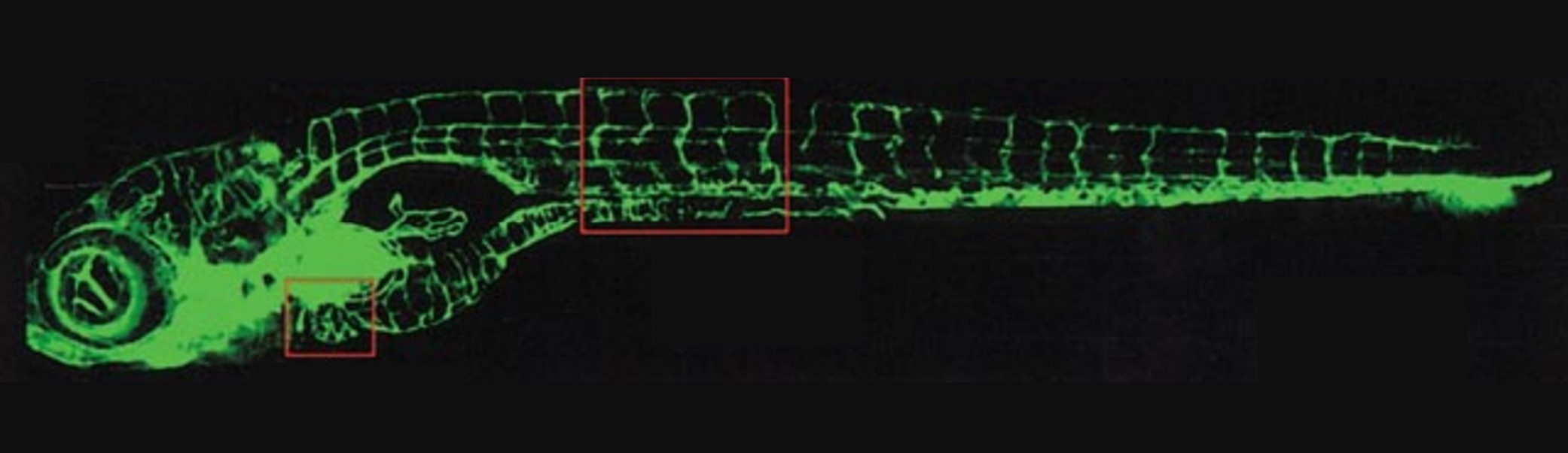
Halloween is just around the corner and zebrafish are in a spooky mood too! Did you know that fish can be fluorescent? Researchers are using fluorescent zebrafish to study a wide range of development mechanisms and pathways.
This article explains how fluorescent zebrafish are obtained and how fluorescence can be detected. It also highlights the areas of use and important discoveries made possible by this technology. Finally, the article also explains how the EggSorter can be adapted to detect fluorescence.
Zebrafish transgenic lines
To obtain fluorescence, zebrafish have to be genetically changed in most cases. That means that a gene coming from other species has to be inserted in the zebrafish DNA (2). Zebrafish are not the only laboratory animals in which an exogenous gene can be inserted. Mice, chickens, xenopus and other fishes such as medaka or killifish can also be fluorescent.
Zebrafish have as some advantages such as the fact that the generation time is short, the embryos develop outside the mother, and that they are transparent so it is possible to observe the development and to track the fluorescence in real time and in vivo (3,4). Moreover, it is easier to target genes in zebrafish than in mice (2).
At this point, it is normal to wonder how an exogenous gene can induce fluorescence. This gene has to contain three different parts (2):
- A promoter which is necessary to indicate the correct orientation of the gene
- A specific gene, often a fluorescent protein gene. These fluorescent proteins possess the unique ability to absorb light at one wavelength and emit light at a different, longer wavelength. Some examples are:
- Blue Fluorescent protein (BFP)
- Green Fluorescent protein (GFP)
- Yellow Fluorescent protein (YFP)
- tdTomato
- mCherry A
- mCherry C

- A termination sequence
The use of fluorescence is very valuable in biology to follow normal biological processes, pathogenic processes or responses to an exposure or intervention (6). So for example, they can be used to explore the expression of a precise protein, by inserting the fluorescence protein gene right after the promoter of the gene for that specific protein of study. In this sense, whenever that gene is active, the fluorescence protein will be expressed and fluorescence will be thus observed.
And how is this gene inserted in the zebrafish genome? There are several methods inducing breaks in DNA that can be employed. Among them, we find :
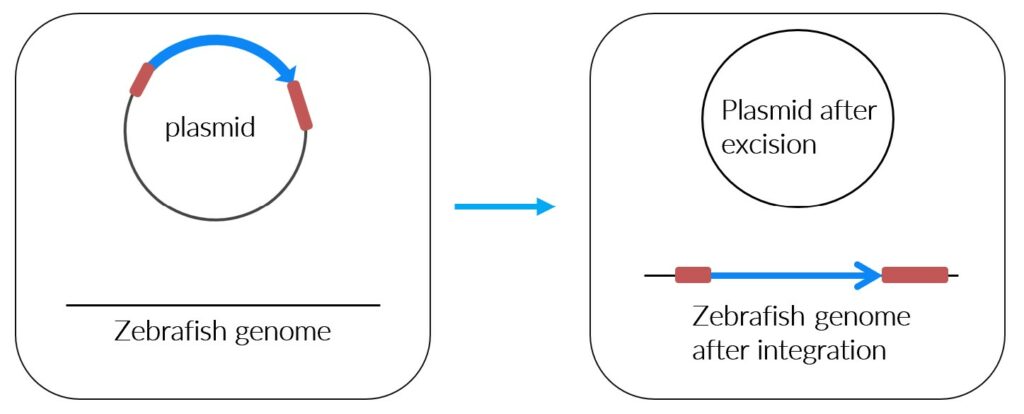
Transposons are genetic elements that can move around within an organism’s DNA. They are divided in copy-and-paste and cut-and-paste mechanisms (7,8). DNA transposons use a cut and paste mechanism while retrotransposons are immobile and move via an RNA intermediate by a mechanism called copy-and-paste. Transposons are inserted into a cell with a plasmid vector.
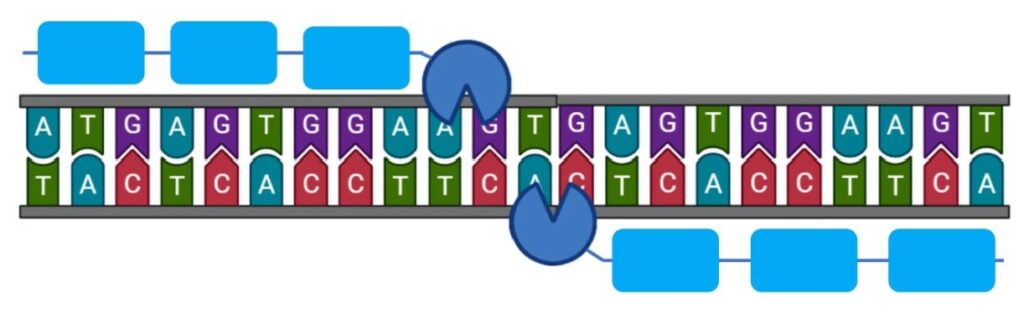
Zinc fingers nuclease induce double strand breaks in a target place in the genome (9) and an exogenous gene can be placed to fill the break.
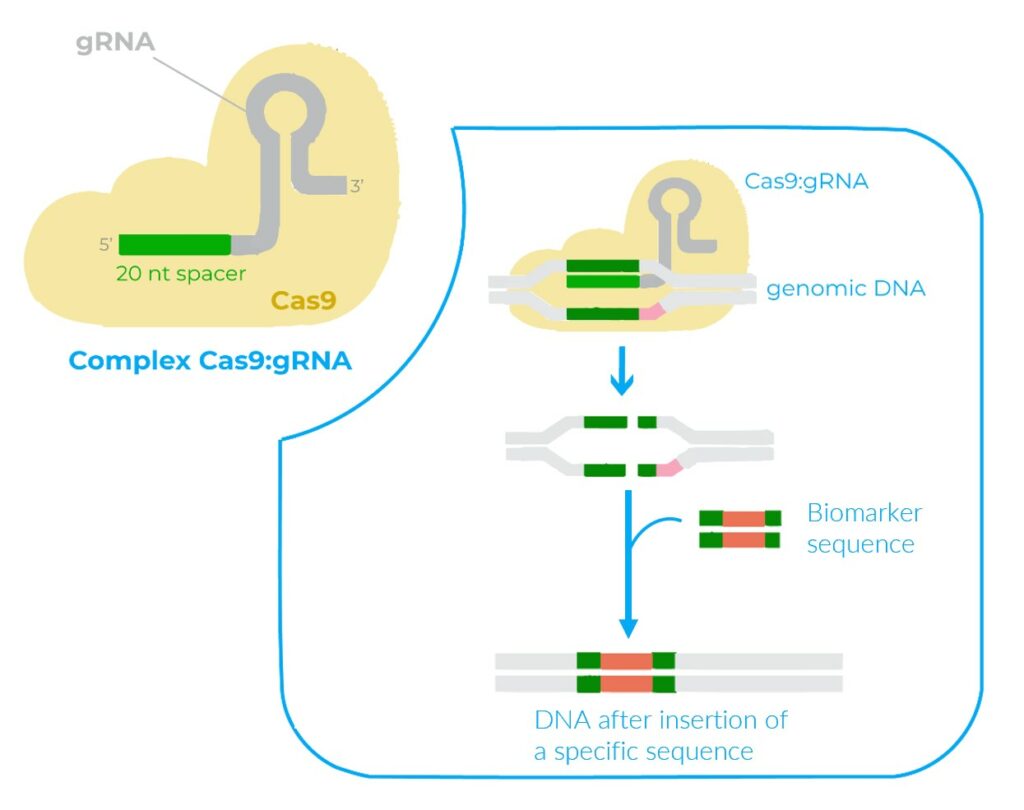
A CRISPR sequence is engineered to be complementary to a specific strand of DNA. The Cas9 enzyme recognizes and cleaves the specific DNA strand using the CRISPR sequence as a guide (10). To learn more about the CRISPR/Cas9 editing system, do not hesitate to read our article “ Precise spatiotemporal genome editing in zebrafish using CRISPR/Cas9” (10).
Fluorescence is also really useful to follow in vivo and in real time gene expression, development mechanisms and different patterns. It is used for exemple to study the regeneration of the nervous system, vasculogenesis and angiogenesis and also to reveal aquatic pollution and ecotoxicology (1,2,4,11). Other systems may also be studied with the establishment of transgenic line and fluorescence, such as the craniofacial skeletal system, the urogenital system or the digestive system (4).
The detection and the visualization of fluorescence cannot be made by naked eyes but they require specialized filters to separate emitted light from excitation light. One major way to visualize it is using fluorescence microscopy. This method allows the visualization at the intracellular scale (12). The fluorescence at a molecular level is obtained by fluorescence spectroscopy and the results are presented in the form of a graph showing intensities at different wavelengths (13).
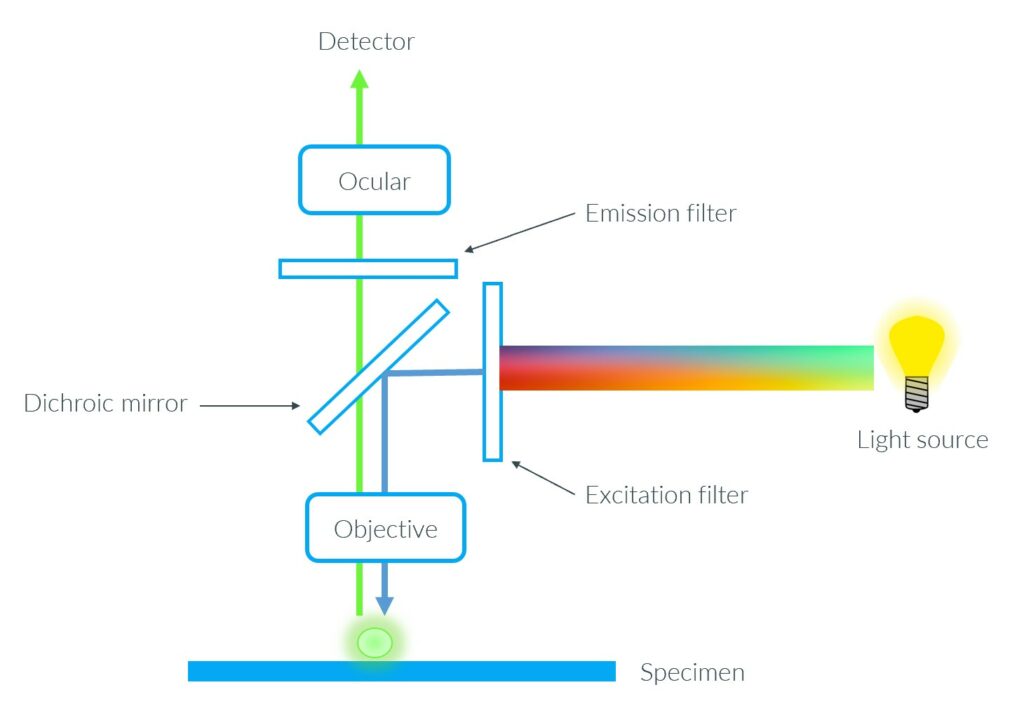
Fluorescence detection in the EggSorter
The EggSorter can detect fluorescence and classify the entities based on the fluorescent expression. Up to six filters are available and they can be used at the same time to allow multi fluorescence.
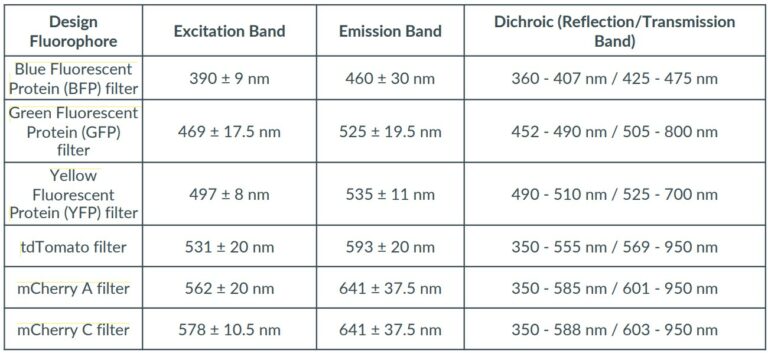
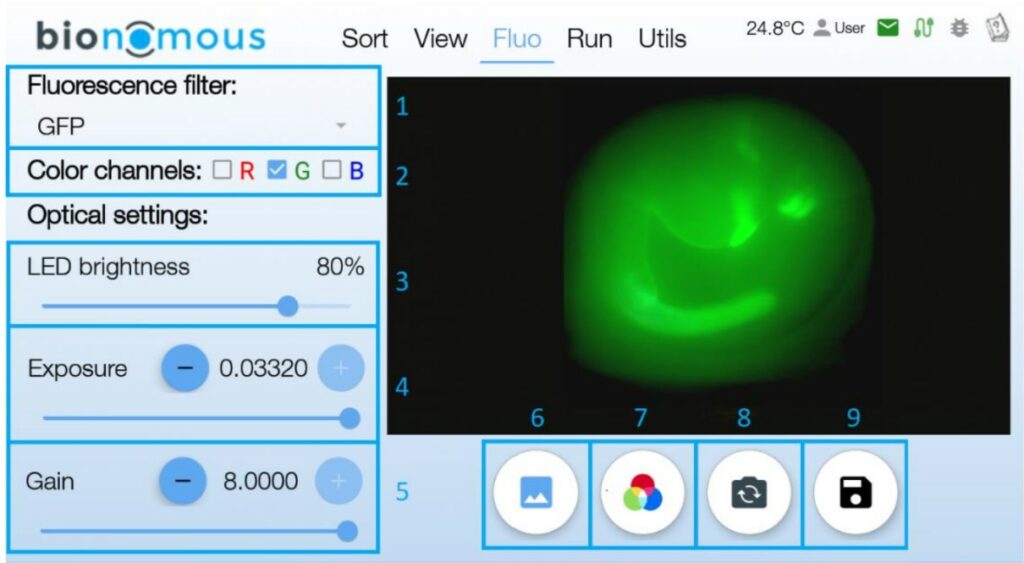
The detection of fluorescence in the EggSorter is by default based on a thresholding mechanism, where the user can set two different thresholds for each filter: an intensity threshold and an area threshold to define what should be considered as a positive fluorescent signal. Besides, our team can create specific algorithms to detect the fluorescence in the most challenging lines. Data can also be extracted from the EggSorter which keeps in memory the fluorescence expression rate for each sample.
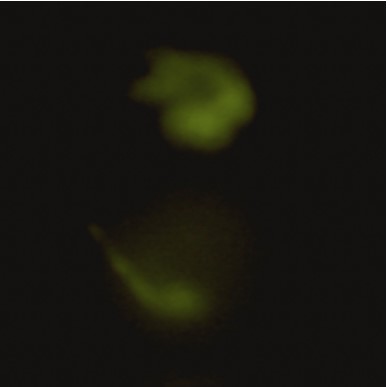
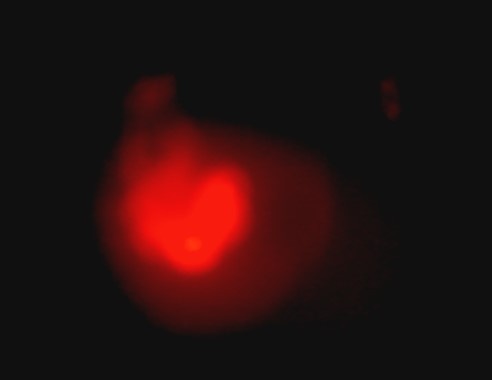
Figure 6: pictures of fluorescent zebrafish embryos obtained by the EggSorter
Conclusion
Whether it is for studying the development of biological systems, monitoring signaling pathways or tracking gene expression, fluorescence is an effective method for real-time in vivo visualization.
The EggSorter can detect multi fluorescence and therefore several mechanisms simultaneously. It is an efficient tool to observe, sort and extract data from transgenic zebrafish embryos.
References
- Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. 2002;
- Brito RS. Transgenic zebrafish (Danio rerio) as an emerging model system in ecotoxicology and toxicology: Historical review, recent advances, and trends. Sci Total Environ. 2022;
- Higashijima S ichi, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-Frequency Generation of Transgenic Zebrafish Which Reliably Express GFP in Whole Muscles or the Whole Body by Using Promoters of Zebrafish Origin. 1997;
- Choe CP. Transgenic fluorescent zebrafish lines that have revolutionized biomedical research. 2021;
- Chakravarthy A, PhD. Fluorescent Proteins in Imaging: Bright and Beautiful. [Internet]. Exploreable. 2011 [cited 2023 Oct 30]. Available from: https://exploreable.wordpress.com/2011/07/28/fluorescent-proteins-in-imaging-bright-and-beautiful/
- Califf RM. Biomarker definitions and their applications. 2018;243.
- Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A, et al. Transposon-mediated genome manipulation in vertebrates. 2009;
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234(2):244–54.
- Third In The Series : Dr. Sharon Amacher – Bionomous [Internet]. [cited 2023 Oct 29]. Available from: https://bionomous.ch/articles/third-in-the-series-sharon-amacher/
- Precise Spatiotemporal Genome Editing In Zebrafish Using CRISPR/Cas9 – Bionomous [Internet]. [cited 2023 Oct 29]. Available from: https://bionomous.ch/articles/spatiotemporal-genome-editing-zebrafish/
- Goldman D, Hankin M, Li Z, Dai X, Ding J. Transgenic Zebrafish for Studying Nervous System Development and Regeneration.
- Sanderson MJ, Smith I, Parker I, Bootman MD. Fluorescence Microscopy. Cold Spring Harb Protoc. 2014 Oct 1;2014(10):pdb.top071795.
- Shirley BA, editor. Protein stability and folding: theory and practice. Totowa, NJ: Humana Press; 1995. 377 p. (Methods in molecular biology).
First image : Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. 2002