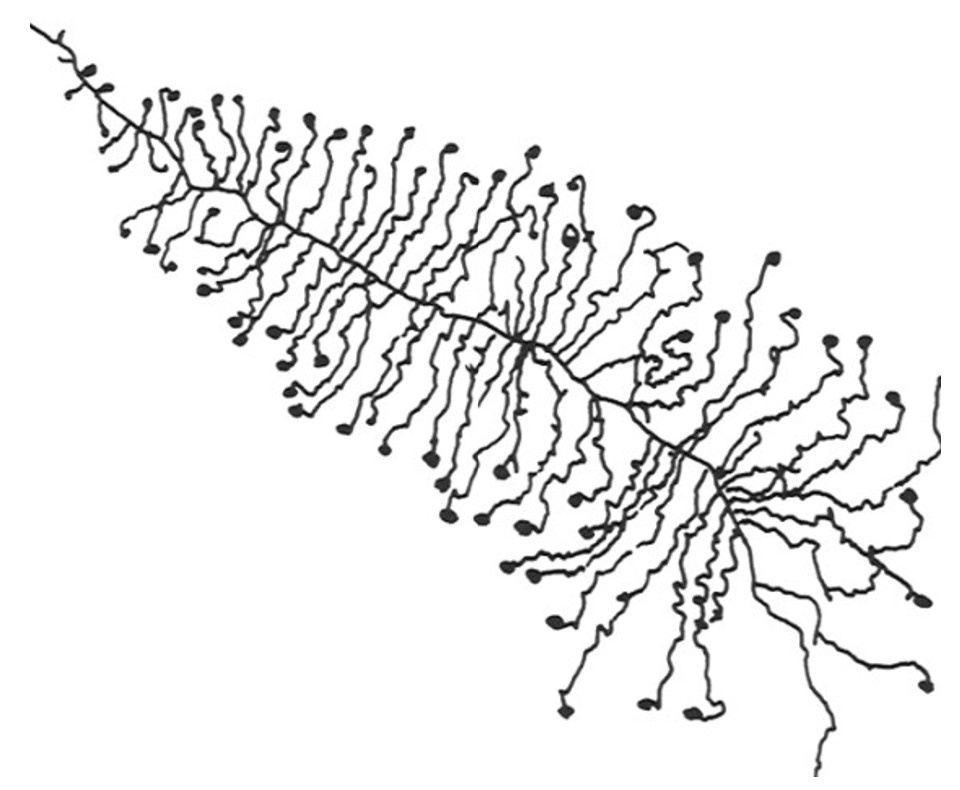
Christmas is coming and even the African clawed frog (Xenopus laevis) cells are in the Christmas spirit. In fact their cells contain a structure that looks like Christmas trees. But what is this particular structure? They are rRNA transcription units in chromatin spreads. And what does this mean? Keep reading to understand it!
In this article we will explain the genetic and transcription bases of this structure.
DNA and RNA
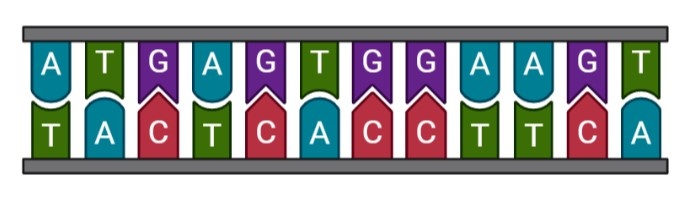
All living organisms are built from deoxyribonucleotide acid (DNA). In animals, most DNA is localized in the nucleus of the cells and contains the genetic instructions used to build proteins. The DNA is organized in a double helix with two chains of nucleotides, which are composed of sugars, phosphate groups and nitrogenous bases. There are four types of nitrogenous base : adenine (A), cytosine (C), guanine (G) and thymine (T). These four nucleotides are combined and the sequence of these bases determine the genetic information. The most important thing is the necessity to have complementation between bases. In fact, A must be in front of T and C must be in front of G. This complementation ensures that no genetic information is lost during transcription.
To go from DNA to proteins, it is necessary to go through an RNA intermediate. RNA is a single chain which looks like DNA but the nitrogenous bases are not exactly the same. In fact, uracil (U) replaces thymine (T). RNA is obtained from DNA by a process called transcription so RNA is a complementary chain of a DNA chain.
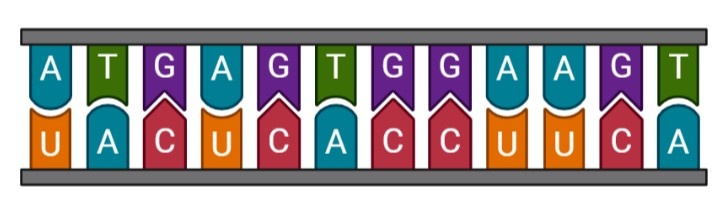
There are different kinds of RNA (see below) but the one which interests us is rRNA. rRNA are transcribed to be an essential component of ribosomes. Ribosomes are the structures in the cells responsible for making proteins.
It also exists mRNA, tRNA and other small RNAs that have regulatory functions in transcription.
Do you want more explanation about mRNA, rRNA and tRNA ? You can find more information by clicking on the RNA names below.
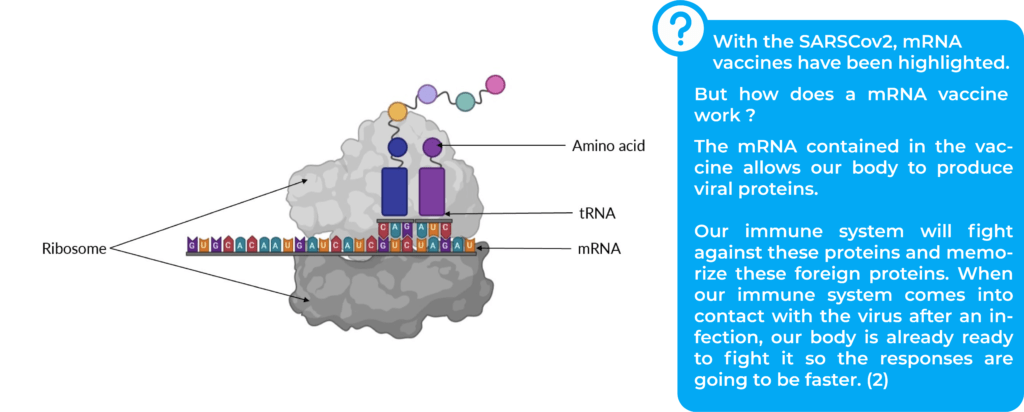
mRNA is a copy of DNA that allows genetic information to leave the cell nucleus. mRNA sequence is read by the ribosome in order to build an amino acid chain specific to each protein (2).
rRNA is synthesized in a special nucleus region. It is an essential component of ribosomes (3).
tRNA finds its complementary codon on mRNA and provides amino acid to constitute protein (4).
Ribosomes read the mRNA to assemble the amino acids that make up the proteins. mRNA is read by codons that are groups with three nitrogenous bases, and the process is called translation.
Christmas tree structure
The particular structure seen in Xenopus laevis is actually present in all eukaryotic cells (1). Eukaryotes represent organisms with DNA contained in a nucleus (5).
The Christmas tree structure takes place in the nucleus and represents active rRNA-coding regions (1,6). Indeed, the trunk of the tree is represented by DNA while each branch is a nascent rRNA (1) and the balls are processing complexes, called 5’ETS process complex (1). This Christmas tree structure is also called “ Miller spreads” in reference to the scientist who visualized this structure for the first time (7).
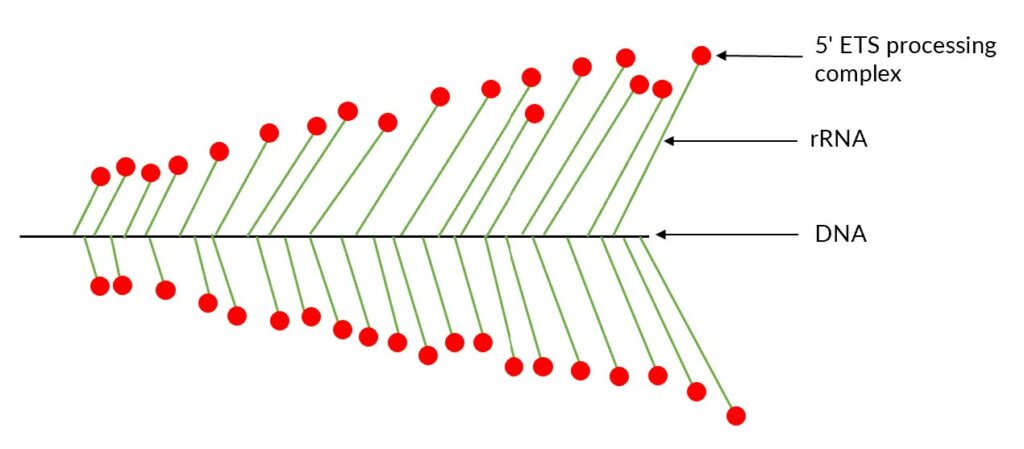
In 1969, Miller and Beatty visualized the Christmas tree structure for the first time in Xenopus laevis. They developed a method still known today as Miller chromatin spreading method.
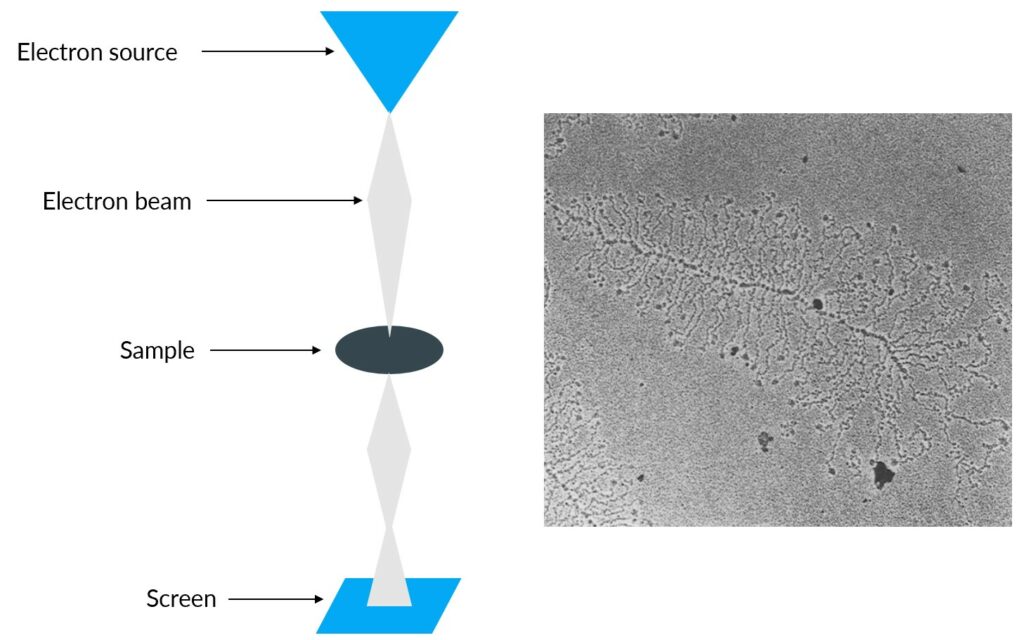
This technique involves the destruction of the cells so that the DNA is released from the nucleus and is dispersed. After centrifugation, the basic nucleosomal structure is conserved (8).They used transmission electron microscopy to see DNA (9). Transmission electron microscopy is used to generate a highly-magnified image (up to 2 million times) (10). Electrons are propelled into the sample and pass through it until they hit a screen at the bottom of the microscope. An image of the sample appears on the screen (10).
Conclusion
DNA and RNA combine to form a Christmas tree structure that was visualized for the first time in 1969 by Miller and collaborators. The techniques they used have greatly helped to advance our knowledge in biology.
The fact that Miller spreads were visualized in Xenopus laevis cells shows its importance as a model organism.
References
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993 Aug;7(8):1609–19.
- Messenger RNA (mRNA) [Internet]. Genome.gov. [cited 2022 Dec 20]. Available from: https://www.genome.gov/genetics-glossary/messenger-rna
- Ribosomal RNA | Definition & Function | Britannica [Internet]. [cited 2022 Dec 20]. Available from: https://www.britannica.com/science/ribosomal-RNA
- Transfer RNA (tRNA) [Internet]. Genome.gov. [cited 2022 Dec 20]. Available from: https://www.genome.gov/genetics-glossary/Transfer-RNA
- eukaryote / eucariote | Learn Science at Scitable [Internet]. [cited 2022 Dec 20]. Available from: https://www.nature.com/scitable/definition/eukaryote-eucariote-294/
- Roger B, Moisand A, Amalric F, Bouvet P. rDNA Transcription during Xenopus laevis Oogenesis. Biochem Biophys Res Commun. 2002 Feb;290(4):1151–60.
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, et al. Pre-18S Ribosomal RNA Is Structurally Compacted into the SSU Processome Prior to Being Cleaved from Nascent Transcripts in Saccharomyces cerevisiae. Mol Cell. 2004 Dec;16(6):943–54.
- Hancock R, editor. The Nucleus: Volume 2: Chromatin, Transcription, Envelope, Proteins, Dynamics, and Imaging [Internet]. Totowa, NJ: Humana Press; 2008 [cited 2022 Dec 22]. (Methods in Molecular Biology; vol. 464). Available from: http://link.springer.com/10.1007/978-1-60327-461-6
- Zannino L, Biggiogera M. How to stain nucleic acids and proteins in Miller spreads. Eur J Histochem EJH. 2022 Feb 25;66(1):3364.
- The Transmission Electron Microscope | CCBER [Internet]. [cited 2022 Dec 22]. Available from: https://www.ccber.ucsb.edu/ucsb-natural-history-collections-botanical-plant-anatomy/transmission-electron-microscope